Note
Click here to download the full example code
Sulcal Identification Query Example in Neurolang¶
Initialise the Neurolang deterministic environment¶
import warnings
warnings.filterwarnings("ignore")
import nibabel as nib
import numpy as np
from matplotlib import pyplot as plt
from nilearn import datasets, plotting
from neurolang import ExplicitVBR, NeurolangDL
Initialise the NeuroLang probabilistic engine.
nl = NeurolangDL()
Load the Destrieux example from nilearn as a fact list
atlas_destrieux = datasets.fetch_atlas_destrieux_2009()
atlas_labels = {
label: str(name.decode("utf8"))
for label, name in atlas_destrieux["labels"]
}
nl.add_atlas_set("destrieux", atlas_labels, nib.load(atlas_destrieux["maps"]))
Out:
destrieux: typing.AbstractSet[typing.Tuple[str, neurolang.regions.ExplicitVBR]] = ('Background', Region(VBR= affine:[[ 2. 0. 0. -76.]
[ 0. 2. 0. -109.]
[ 0. 0. 2. -64.]
[ 0. 0. 0. 1.]], voxels:[[ 0 0 0]
[ 0 0 1]
[ 0 0 2]
...
[75 92 73]
[75 92 74]
[75 92 75]])) ...
Add utility functions, one for the prefix of the region’s name one to determine the principal direction of the region.
@nl.add_symbol
def startswith(prefix: str, s: str) -> bool:
"""Describe the prefix of string `s`.
Parameters
----------
prefix : str
prefix to query.
s : str
string to check whether its
prefixed by `s`.
Returns
-------
bool
whether `s` is prefixed by
`prefix`.
"""
return s.startswith(prefix)
@nl.add_symbol
def principal_direction(s: ExplicitVBR, direction: str, eps=1e-6) -> bool:
"""Describe the principal direction of
the extension of a volumetric region.
Parameters
----------
s : ExplicitVBR
region to analyse the principal
direction of its extension.
direction : str
principal directions, one of
`LR`, `AP`, `SI`, for the directions
left-right, anterio-posterior, and
superior inferior respectively.
eps : float, optional
minimum difference on between
directional standard deviations,
by default 1e-6.
Returns
-------
bool
wether the principal variance of
`s` is `direction`.
"""
# Assuming RAS coding os the xyz space.
c = ["LR", "AP", "SI"]
s_xyz = s.to_xyz()
cov = np.cov(s_xyz.T)
evals, evecs = np.linalg.eig(cov)
i = np.argmax(np.abs(evals))
abs_max_evec = np.abs(evecs[:, i].squeeze())
sort_dir = np.argsort(abs_max_evec)
if np.abs(abs_max_evec[sort_dir[-1]] - abs_max_evec[sort_dir[-2]]) < eps:
return False
else:
main_dir = c[sort_dir[-1]]
return (direction == main_dir) or (direction[::-1] == main_dir)
Example 1: Characterise Some of the Sulci¶
In this example we characterise:
left hemisphere primary sulci, by name
left frontal lobe sulcus as those
anterior to Destrieux’s left central sulcus
superior to Destrieux’s left anterio-vertical section of the lateral fissure.
These will be present in all further programs. There are no executed queries in this section, just declared ones.
with nl.environment as e:
e.left_sulcus[e.name_, e.region] = e.destrieux(
e.name_, e.region
) & startswith("L S", e.name_)
e.left_primary_sulcus[e.name_, e.region] = e.destrieux(e.name_, e.region) & (
(e.name_ == "L S_central")
| (e.name_ == "L Lat_Fis-post")
| (e.name_ == "L S_pericallosal")
| (e.name_ == "L S_parieto_occipital")
| (e.name_ == "L S_calcarine")
| (e.name_ == "L Lat_Fis-ant-Vertical")
| (e.name_ == "L Lat_Fis-ant-Horizont")
)
e.left_frontal_lobe_sulcus[e.region] = (
e.left_sulcus(..., e.region)
& e.anatomical_anterior_of(e.region, e.destrieux.s["L S_central"])
& e.anatomical_superior_of(
e.region, e.destrieux.s["L Lat_Fis-ant-Vertical"]
)
)
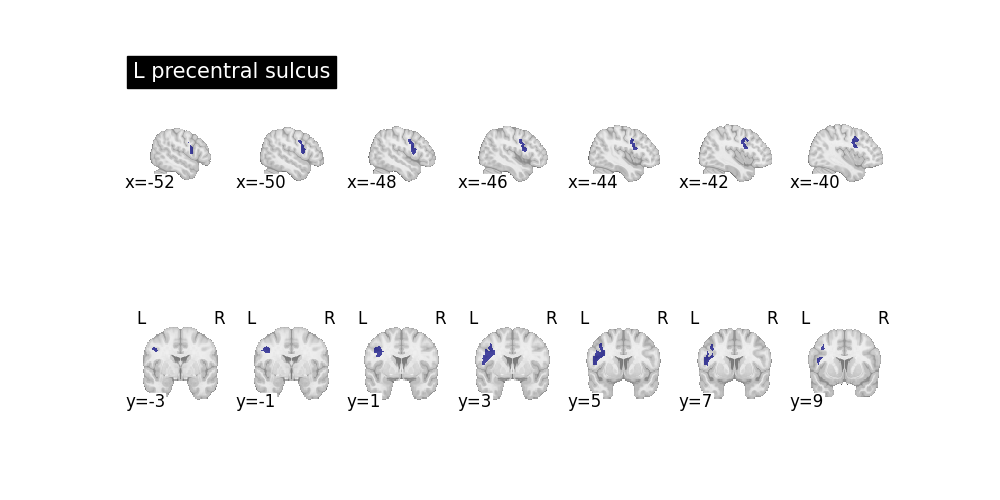
Example 2: Query the Precentral Sulcus¶
this query and all defined will not be in the program after the with context finishes. But the results remain. We identify the precentral sulcus (PC) as:
belongs to the left frontal lobe
its principal direction is along the superior-inferior axis.
no other sulcus satisfying the same conditions is anterior to the PC.
with nl.scope as e:
e.named_sulcus["L precentral sulcus", e.region] = (
e.left_frontal_lobe_sulcus(e.region)
& e.principal_direction(e.region, "SI")
& ~nl.exists(
e.other_region,
e.left_frontal_lobe_sulcus(e.other_region)
& (e.region != e.other_region)
& e.anatomical_posterior_of(e.other_region, e.region),
)
)
res = nl.query((e.name_, e.region), e.named_sulcus(e.name_, e.region))
for name, region in res:
subplots = plt.subplots(nrows=2, ncols=1, figsize=(10, 5))[1]
plotting.plot_roi(
region.spatial_image(), display_mode="x", title=name, axes=subplots[0]
)
plotting.plot_roi(
region.spatial_image(), display_mode="y", axes=subplots[1]
)
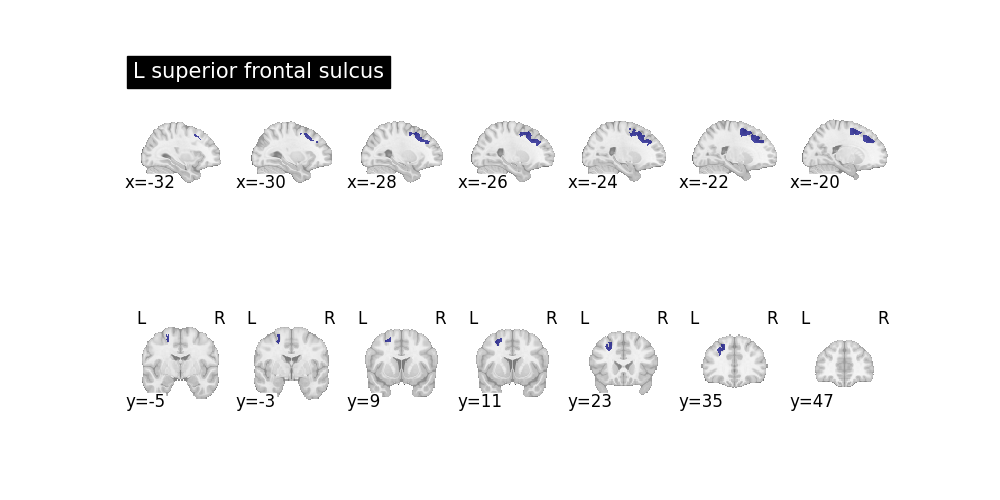
Example 3: Query the Superior Frontal Sulcus¶
this query and all defined will not be in the program after the with context finishes. But the results remain. In this query we express that the superior frontal sulcus (SFS) as a sulcus which:
belongs to the left frontal lobe
its principal direction is along the anterior-posterior axis.
no other sulcus satisfying the same conditions is superior to the SFS.
with nl.scope as e:
e.named_sulcus["L superior frontal sulcus", e.region] = (
e.left_frontal_lobe_sulcus(e.region)
& e.principal_direction(e.region, "AP")
& ~nl.exists(
e.other_region,
e.left_frontal_lobe_sulcus(e.other_region)
& (e.region != e.other_region)
& e.anatomical_superior_of(e.other_region, e.region),
)
)
res = nl.query((e.name_, e.region), e.named_sulcus(e.name_, e.region))
for name, region in res:
subplots = plt.subplots(nrows=2, ncols=1, figsize=(10, 5))[1]
plotting.plot_roi(
region.spatial_image(), display_mode="x", title=name, axes=subplots[0]
)
plotting.plot_roi(
region.spatial_image(), display_mode="y", axes=subplots[1]
)
Total running time of the script: ( 0 minutes 14.061 seconds)
