Note
Go to the end to download the full example code
NeuroLang Example based Implementing a NeuroSynth Query¶
import warnings
warnings.filterwarnings("ignore")
from pathlib import Path
from typing import Iterable
import nibabel
import nilearn.datasets
import nilearn.image
import nilearn.plotting
import numpy as np
import pandas as pd
from neurolang import ExplicitVBROverlay, NeurolangPDL
from neurolang.frontend.neurosynth_utils import get_ns_mni_peaks_reported
Data preparation¶
data_dir = Path.home() / "neurolang_data"
Load the MNI atlas and resample it to 4mm voxels
mni_t1 = nibabel.load(
nilearn.datasets.fetch_icbm152_2009(data_dir=str(data_dir / "icbm"))["t1"]
)
mni_t1_4mm = nilearn.image.resample_img(mni_t1, np.eye(3) * 4)
Dataset created in /home/circleci/neurolang_data/icbm/icbm152_2009
Downloading data from https://osf.io/7pj92/download ...
...done. (2 seconds, 0 min)
Extracting data from /home/circleci/neurolang_data/icbm/icbm152_2009/e05b733c275cab0eec856067143c9dc9/download..... done.
Probabilistic Logic Programming in NeuroLang¶
nl = NeurolangPDL()
Adding new aggregation function to build a region overlay
@nl.add_symbol
def agg_create_region_overlay(
i: Iterable, j: Iterable, k: Iterable, p: Iterable
) -> ExplicitVBROverlay:
mni_coords = np.c_[i, j, k]
return ExplicitVBROverlay(
mni_coords, mni_t1_4mm.affine, p, image_dim=mni_t1_4mm.shape
)
Load the NeuroSynth database
peak_data = get_ns_mni_peaks_reported(data_dir)
ijk_positions = np.round(
nibabel.affines.apply_affine(
np.linalg.inv(mni_t1_4mm.affine),
peak_data[["x", "y", "z"]].values.astype(float),
)
).astype(int)
peak_data["i"] = ijk_positions[:, 0]
peak_data["j"] = ijk_positions[:, 1]
peak_data["k"] = ijk_positions[:, 2]
peak_data = peak_data[["i", "j", "k", "id"]]
nl.add_tuple_set(peak_data, name="PeakReported")
study_ids = nl.load_neurosynth_study_ids(data_dir, "Study")
nl.add_uniform_probabilistic_choice_over_set(
study_ids.value, name="SelectedStudy"
)
nl.load_neurosynth_term_study_associations(
data_dir, "TermInStudyTFIDF", tfidf_threshold=1e-3
)
Downloading data from https://github.com/neurosynth/neurosynth-data/raw/master/data-neurosynth_version-7_coordinates.tsv.gz ...
...done. (1 seconds, 0 min)
Downloading data from https://github.com/neurosynth/neurosynth-data/raw/master/data-neurosynth_version-7_metadata.tsv.gz ...
...done. (0 seconds, 0 min)
Downloading data from https://github.com/neurosynth/neurosynth-data/raw/master/data-neurosynth_version-7_vocab-terms_source-abstract_type-tfidf_features.npz ...
...done. (0 seconds, 0 min)
Downloading data from https://github.com/neurosynth/neurosynth-data/raw/master/data-neurosynth_version-7_vocab-terms_vocabulary.txt ...
...done. (0 seconds, 0 min)
TermInStudyTFIDF: typing.AbstractSet[typing.Tuple[str, str, float]] = (9862924, '001', 0.0553942161114) ...
with nl.scope as e:
e.Activation[e.i, e.j, e.k] = e.PeakReported(
e.i, e.j, e.k, e.s
) & e.SelectedStudy(e.s)
e.TermAssociation[e.t] = e.SelectedStudy(e.s) & e.TermInStudyTFIDF(
e.s, e.t, ...
)
e.ActivationGivenTerm[e.i, e.j, e.k, e.PROB[e.i, e.j, e.k]] = e.Activation(
e.i, e.j, e.k
) // e.TermAssociation("auditory")
e.ActivationGivenTermImage[
agg_create_region_overlay(e.i, e.j, e.k, e.p)
] = e.ActivationGivenTerm(e.i, e.j, e.k, e.p)
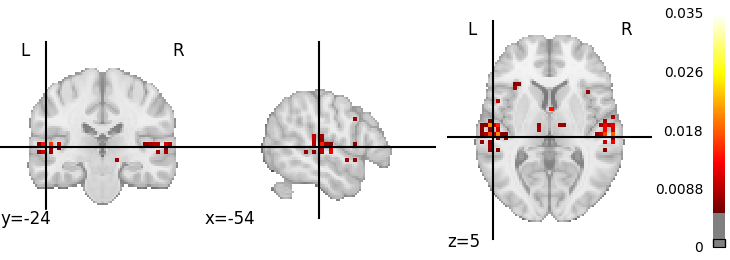
img_query = nl.query((e.x,), e.ActivationGivenTermImage(e.x))
result_image = img_query.fetch_one()[0].spatial_image()
img = result_image.get_fdata()
plot = nilearn.plotting.plot_stat_map(
result_image, threshold=np.percentile(img[img > 0], 95)
)
nilearn.plotting.show()
Total running time of the script: (0 minutes 12.899 seconds)